Chapter 4 Molecular Basis of Inheritance
1. Multiple Choice Questions
Question 1.
Griffith worked on ………………..
(a) Bacteriophage
(b) Drosophila
(c) Frog eggs
(d) Streptococci
Answer:
(d) Streptococci
Question 2.
The molecular knives of DNA are ………………..
(a) Ligases
(b) Polymerases
(c) Endonucleases
(d) Transcriptase
Answer:
(c) Endonucleases
Question 3.
Translation occurs in the ………………..
(a) Nucleus
(b) Cytoplasm
(c) Nucleolus
(d) Lysosomes
Answer:
(b) Cytoplasm
Question 4.
The enzyme required for transcription is ………………..
(a) DNA polymerase
(b) RNA polymerase
(c) Restriction enzyme
(d) RNase
Answer:
(b) RNA polymerase
Question 5.
Transcription is the transfer of genetic information from ………………..
(a) DNA to RNA
(b) t-RNA to m-RNA
(c) DNA to m-RNA
(d) m-RNA to t-RNA
Answer:
(a) DNA to RNA
Question 6.
Which of the following is NOT part of protein synthesis?
(a) Replication
(b) Translation
(c) Transcription
(d) All of these
Answer:
(a) Replication
Question 7.
In the RNA molecule, which nitrogen base is found in place of thymine?
(a) Guanine
(b) Cytosine
(c) Thymine
(d) Uracil
Answer:
(d) Uracil
Question 8.
How many codons are needed to specify three amino acids?
(a) 3
(b) 6
(c) 9
(d) 12
Answer:
(a) 3
Question 9.
Which out of the following is NOT an example of inducible operon?
(a) Lactose operon
(b) Histidine operon
(c) Arabinose operon
(d) Tryptophan operon
Answer:
(d) Tryptophan operon
Question 10.
Place the following event of translation in the correct sequence ………………..
i. Binding of met-t-RNA to the start codon.
ii. Covalent bonding between two amino acids.
iii. Binding of second t-RNA.
iv. Joining of small and large ribosome subunits.
(a) iii, iv, i, ii
(b) i, iv, iii, ii
(c) iv, iii, ii, i
(d) ii, iii, iv, i
Answer:
(b) i, iv, iii, ii
2. Very Short Answer Questions
Question 1.
What is the function of an RNA primer during protein synthesis?
Answer:
During DNA replication, RNA primer provides 3’ OH to which DNA polymerase enzyme can add nucleotides to synthesize new strand using parental strand of DNA as template.
[Note : RNA primer has no direct role in protein synthesis.]
Question 2.
Why is the genetic code considered as commaless?
Answer:
The triplet codon are arranged one after the other on m-RNA molecule without any gap or space and therefore genetic code is considered as commaless.
Question 3
Genome
Answer:
Genome is the total genetic constitution of an organism or a complete copy of genetic information (DNA) or one complete set of chromosomes (monoploid or haploid) of an organism.
Question 4.
Which enzyme does remove supercoils from replicating DNA?
Answer:
Super-helix relaxing enzyme (Topoisomerase) removes supercoils from replicating DNA.
Question 5.
Why are Okazaki fragments formed on lagging strand only?
Answer:
Okazaki fragments are formed only on lagging template as only short stretch of lagging template becomes available for replication at one time.
Question 6.
When does DNA replication take place?
Answer:
In eukaryotes DNA-replication takes place during S-phase of interphase of cell cycle and in prokaryotes. DNA replicates prior to cell division.
Question 7.
Define term Codogen and Codon
Answer:
Codogen is a triplet of nucleotides present on the DNA which specifies one particular amino acid.
Codon is a triplet of nucleotides present on the m-RNA which specifies one particular amino acid.
Question 8.
What is degeneracy of genetic code?
Answer:
Genetic code is degenerate as 61 codons code for 20 amino acids, that is two or more codons can specify the same amino acid. E.g. Cysteine has two codons, while isoleucine has three codons.
Question 9.
Which are the nucleosomal ‘core’ histones?
Answer:
Two molecules each of histone proteins, viz. H2A. H2B, H3 and H4 are the nucleosomal ‘core’ histones.
3. Short Answer Questions
Question 1.
DNA packaging in eukaryotic cell.
Answer:
- In eukaryotic cells, DNA (2.2 metres) is condensed, coiled and supercoiled to be packaged efficiently in the nucleus (10-16 m).
- DNA is associated with histone and non-histone proteins.
- Histones are a set of positively charged, basic proteins, rich in basic amino acid residues lysine and arginine.
- Nucleosome consists of nucleosome core (two molecules of each of histone proteins viz. H2A, H2B, H3 and H4 forming histone octamer) and negatively charged DNA (146 bps) that wraps around the histone octamer by 1 3/4 turns.
- H1 protein binds the DNA thread where it enters and leaves the nucleosome.
- Adjacent nucleosomes are linked with linker DNA (varies in length from 8 to 114 bp, average length of linker DNA is about 54 bp).
- Each nucleosome contains 200 bp of DNA.
- Packaging involves formation of – Beads on string (10 nm diameter), Solenoid fibre (looks like coiled telephone wire, 30 nm diameter/300Å), Chromatin fibre and Chromosome.
- Non-Histone Chromosomal Proteins (NHC) contribute to the packaging of chromatin at a higher level.
Question 2.
Enlist the characteristics of genetic code.
Answer:
The characteristics of genetic code are
- Genetic code is triplet, commaless and non-overlapping.
- It is degenerate and non-ambiguous.
- It is universal
- It has polarity.
Question 3.
Applications of DNA fingerprinting.
Answer:
Applications of DNA fingerprinting are as follows:
- In forensic science to solve rape and murder cases.
- Finds out the biological father or mother or both, of the child, in case of disputed parentage.
- Used in pedigree analysis in cats, dogs, horses and humans.
Question 4.
Explain the role of lactose in ‘Lac Operon’.
Answer:
- A small amount of beta-galactoside permease enzyme is present in cell even when Lac operon is switched off and it allows a few molecules of lactose to enter into the cell.
- Lactose binds to repressor and inactivates it.
- Repressor – lactose complex cannot bind with the operator gene, which is then turned on.
- RNA polymerase transcribes all the structural genes to produce lac m-RNA which is then translated to produce all enzymes.
- Thus, lactose acts as an inducer.
- When the inducer level falls, the operator is blocked again by repressor and structural genes are repressed again. This is negative feedback.
4. Short Answer Questions
Question 1.
Human genome project.
Answer:
1. Human Genome Project (HGP) was initiated in 1990 under the International administration of the Human Genome Organization (HUGO) and it was completed r in 2003.
2. The main aims:
- To sequence 3 billion base pairs of DNA in human genome and to map an estimated 33000 genes.
- To store the information collected from the project in databases.
- To develop tools and techniques for analysis of the data.
- Transfer of the related technologies to the private sectors, such as industries.
- Taking care of the legal, ethical and social issues which may arise from project.
- To sequence the genomes of several other organisms such as bacteria e.g. E.coli, Caenorhabditis elegans, Saccharomyces cerevisiae, Drosophil, rice, Arabidopsis), Mus musculus, etc.
3. Significance:
- HGP has a major impact in the fields like Medicine, Biotechnology, Bioinformatics and the Life sciences.
- More understanding of functions of genes, proteins and human evolution.
Question 2.
Describe the structure of operon.
Answer:
- An operon is a unit of gene expression and regulation.
- It includes the structural genes and their control elements. Control elements are promoters and operators.
- The structural genes code for proteins, r-RNA and t-RNA that are necessary for all the cells.
- Promoters are signal sequences in DNA. They start the RNA synthesis. They also act as sites where the RNA polymerases are bound during transcription.
- Operators are present between the promoters and structural genes.
- There is repressor protein that binds to the operator region of the operon.
- There are regulatory genes which are responsible for the formation of repressors which interact with operators.
Question 3.
In the figure below A, B and C are three types of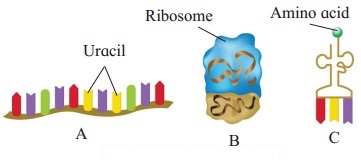
Answer:
Answer: A, B and C are A : m-RNA, B : r-RNA, C : t-RNA
Question 4.
Identify the labelled structures on the following diagram of translation.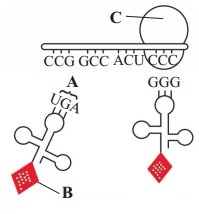
Part A is the ………………
Part B is the ………………
Part C is the ………………
Answer:
Part A is the anti-codon.
Part B is the amino acid.
Part C is the larger subunit of ribosome.
Question 5.
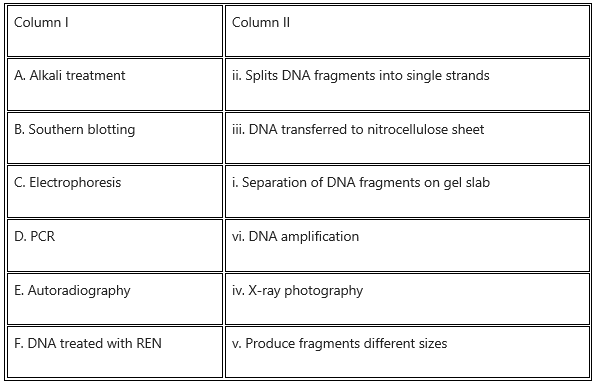
Match the entries in Column I with those of Column II and choose the correct answer.
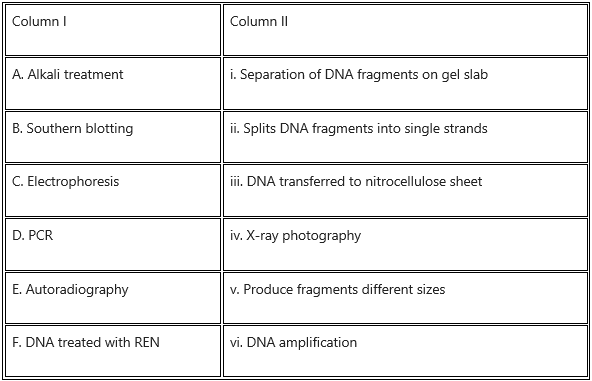
Answer:
5. Long Answer Questions
Question 1.
Explain the process of DNA replication.
Answer:
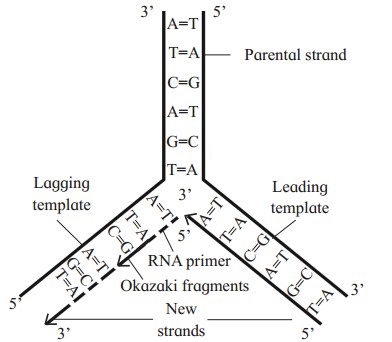
DNA replication is semi-conservative replication. It involves following steps:
Activation of Nucleotides:
- Nucleotides (dAMP dGMR dCMP and dTMP) present in the nucleoplasm, are activated by ATP in presence of an enzyme phosphorylase.
- This phosphorylation results in the formation of deoxyribonucleotide triphosphates i.e. dATE dGTR dCTP and dTTE
Point of Origin or Initiation point:
- Replication begins at specific point ‘O- Origin and terminates at point ‘T’.
- At the point ‘O’, enzyme endonuclease nicks (breaks the sugar-phosphate backbone or the phosphodiester bond) one of the strands of DNA, temporarily.
Unwinding of DNA molecule:
- Enzyme DNA helices breaks weak hydrogen bonds in the vicinity of ‘O’.
- The strands of DNA separate and unwind. This unwinding is bidirectional.
- SSBP (Single strand binding proteins) remains attached to both the separated strands and prevent them from recoiling (rejoining).
Replicating fork:
- Y-shape replication fork is formed due to unwinding and separation of two strands.
- The unwinding of strands results in strain which is released by super-helix relaxing enzyme.
Synthesis of new strands:
- Each separated strand acts as a template for the synthesis of new complementary strand.
- A small RNA primer (synthesized by activity of enzyme RNA primase) get attached to the 3′ end of template strand and attracts complementary nucleotides from surrounding nucleoplasm.
- These nucleotides bind to the complementary nucleotides on the template strand by hydrogen bonds (i.e. A = T or T = A; G = C or C = G, CEG).
- The phosphodiester bonds are formed between nucleotides of new strand to form a polynucleotide strand.
- The enzyme DNA polymerase catalyses synthesis of new complementary strand always in 5′ – 3′ direction.
Leading and Lagging strand:
- The template strand with free 3′ is called the leading template.
- The template strand with free 5′ end is called the lagging template.
- The replication always starts at C-3 end of template strand and proceeds towards C-5 end.
- New strands are always formed in 5′ → 3′ direction.
- The new strand which develops continuously towards replicating fork is called the leading strand.
- The new strand which develops discontinuously away from the replicating fork is called the lagging strand.
- Maturation of Okazaki fragments : The lagging strand is synthesized in the form of small Okazaki fragments which are joined by enzyme DNA ligase.
- Later RNA primers are removed by the combined action of RNase H, an enzyme that degrades the RNA strand of RNA-DNA hybrids, and polymerase I.
- Gaps formed are filled by complementary DNA sequence with the help of DNA polymerase-I in prokaryotes and DNA polymerase-a in eukaryotes.
- Finally, DNA gyrase (topoisomerase) enzyme forms double helix to form daughter DNA molecules.
Formation of two daughter DNA molecules:
- In each daughter DNA molecule, one strand is parental and the other one is newly synthesized.
- Thus, 50% part (i.e. one strand of the helix) is contributed by mother DNA. Hence, it is described as semiconservative replication.
Question 2.
Describe the process of transcription in protein synthesis.
Answer:
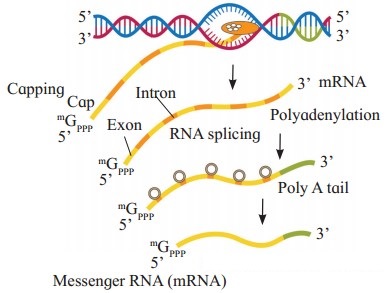
Transcription involves three stages, viz. Initiation, Elongation and Termination.
(1) Initiation:
- RNA polymerase binds to promoter site.
- It then moves along the DNA and causes local unwinding of DNA duplex into two strands in the region of the gene.
- Only antisense strand functions as template.
(2) Elongation:
- The complementary ribonucleoside tri-phosphates get attached to exposed bases of DNA template chain.
- As transcription proceeds, the hybrid DNA-RNA molecule dissociates and makes m-RNA molecule free.
- As the m-RNA grows, the transcribed region of DNA molecule becomes spirally coiled and regains double helical form.
(3) Termination:
When RNA polymerase reaches the terminator site on the DNA, both enzyme and newly formed m-RNA (primary transcript) gets released.
Question 3.
Describe the process of translation in protein synthesis.
Answer:
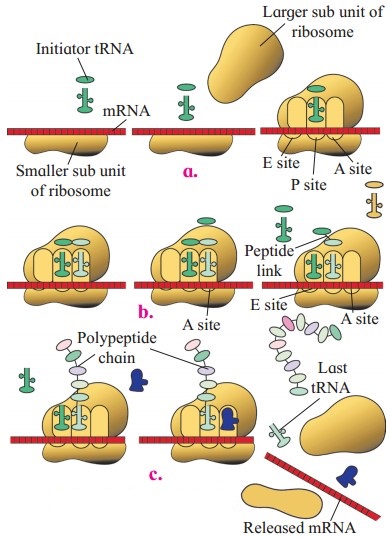
Translation involves the following steps:
1. Activation of amino acids and formation of charged t-RNA (t-RNA – amino acid complex):
i. In the presence of an enzyme amino acyl t-RNA synthetase, the amino acid is activated and then attached to the specific t-RNA molecule at 3’ end to form charged t-RNA (t-RNA – amino acid complex).
ii. ATP is essential for the reaction.
2. Initiation of Polypeptide chain:
- Small subunit of ribosome binds to the m-RNA at 5’ end.
- Start codon is positioned properly at P-site.
- Initiator t-RNA, (carrying amino acid methionine in eukaryotes or formyl methionine in prokaryotes) binds with initiation codon (AUG) of m-RNA, by its anticodon (UAC) through hydrogen bonds.
- The large subunit of ribosome joins with the smaller subunit in the presence of Mg++.
- Thus, initiator charged t-RNA occupies the P-site and A – site is vacant.
3. Elongations of polypeptide chain:
Addition of amino acid occurs in 3 Step cycle-
i. Codon recognition.
Anticodon of second (and subsequent) amino acyl t-RNA molecule recognizes and binds with codon at A-site by hydrogen bonds.
ii. Peptide bond formation.
- Ribozyme catalyzes the peptide bond formation between amino acids on the initiator t-RNA at P-site and t-RNA at A-site.
- It takes less than 0.1 second for formation of peptide bond.
- Initiator t-RNA at ‘P’ site is then released from E-site.
iii. Translocation.
- Translocation is the process in which sequence of codons on m-RNA is decoded and accordingly amino acids are added in specific sequence to form a polypeptide on ribosomes.
- Due to this A’-site becomes vacant to receive next charged t-RNA molecule.
- The events like arrival of t-RNA – amino acid complex, formation of peptide bond, ribosomal translocation and release of previous t-RNA, are repeated.
- As ribosome move over the m-RNA, all the codons on m-RNA are exposed oiie by one for translation.
4. Termination and release of polypeptide:
When stop codon (UAA, UAG, UGA) gets exposed at the A-site, the release factor binds to the stop codon, thereby terminating the translation process
The polypeptide gets released in the cytoplasm.
Two subunits of ribosome dissociate and last t-RNA and m-RNA are released in the cytoplasm.
m-RNA gets denatured by nucleases immediately.
Question 4.
Describe Lac ‘Operon’.
Answer:
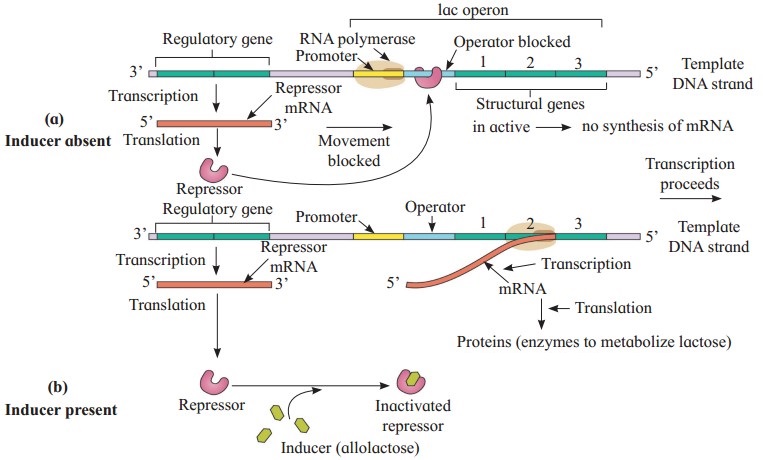
Lac operon consists of the following components:
(1) Regulator gene:
- Regulator gene precedes the promoter gene.
- It may not be present immediately adjacent to operator gene.
- Regulator gene codes for a repressor protein which binds with operator gene and represses (stops) its action.
(2) Promoter gene:
- It precedes the operator gene.
- It is present adjacent to operator gene.
- RNA Polymerase enzyme binds at promoter site.
- Promoter gene base sequence determines which strand of DNA acts a template.
(3) Operator gene:
- It precedes the structural genes.
- When operator gene is turned on by an inducer, the structural genes get transcribed to form m-RNA.
(4) Structural gene:
- There are 3 structural genes in the sequence lac-Z, lac-Y and lac-A.
-
Enzymes produced are β-galactosidase, β-galactoside permease and transacetylase respectively.
Inducer Allolactose acts as an inducer. It inactivates the repressor by binding with it.
Question 5.
Justify the statements. If the answer is false, change the underlined word(s) to make the statement true.
(i) The DNA molecule is double stranded and the RNA molecule is single stranded.
Answer:
- DNA as the genetic material has to be chemically and structurally stable.
- It should be able to generate its replica.
- Sugar-phosphate backbone and complementary base pairing between the two strands, give stability to DNA.
- Both the strands of DNA act as template for synthesis of their complementary strands. This allows accurate replication of DNA.
- Single stranded RNA can be folded to form complex structures and perform specific functions such as synthesis of proteins.
(ii) The process of translation occurs at the ribosome.
Answer:
- Translation is the process in which sequence of codons of m-RNA is decoded and accordingly amino acids are added in specific sequence to form a polypeptide on ribosomes.
- Ribosome has one binding site for m-RNA. It orients m-RNA molecule in such a way that all the codons are properly read.
- Ribosome has three binding sites for t-RNA : P-site (peptidyl t-RNA-site), A-site (aminoacyl t-RNA-site) and E-site (exit site).
- t-RNAs place the required amino acids in correct sequence and translate the coded message of RNA.
- In eukaryotes, a groove which is present between two subunits of ribosomes, protects the polypeptide chain from the action of cellular enzymes and also protects m-RNA from the action of nucleases.
- Thus ribosome plays an important role in translation.
(iii) The job of m-RNA is to pick up amino acids and transport them to the ribosomes.
Answer:
The job of t-RNA is to pick up amino acids and transport them to ribosomes. t-RNA is an adapter molecule. It reads the codons of m-RNA and also simultaneously transfer specific amino acid to m-RNA Ribosome complex. It binds with amino acid at its 3′ end.
(iv) Transcription must occur before translation may occur.
Answer:
In prokaryotes, translation can start before transcription is complete, as both these processes occur in the same compartment, i.e. cytoplasm. But in eukaryotes, transcription and processing of hnRNA occurs in nucleus. hnRNA then comes out of the nucleus through nuclear pores and then it is translated at ribosomes in the cytoplasm.
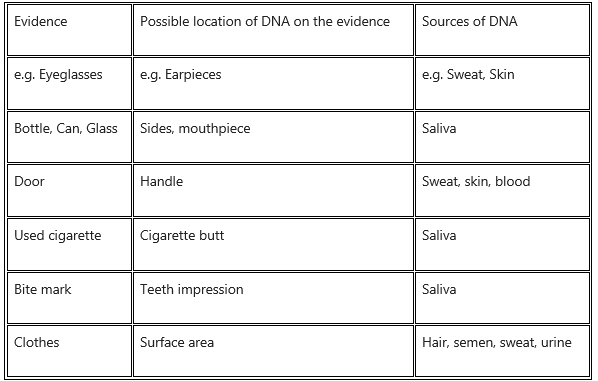
Question 6.
Guess
(i) the possible locations of DNA on the collected evidence from a crime scene and
(ii) the possible sources of DNA.
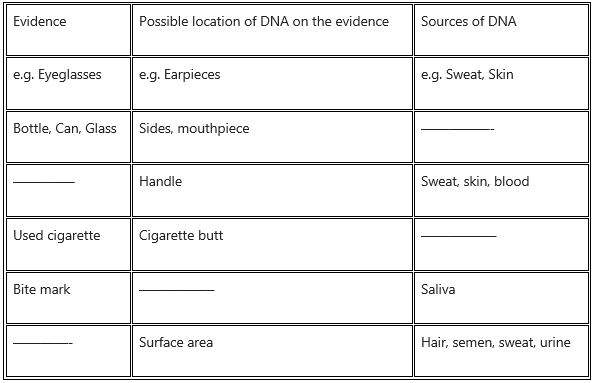
Answer: